Screening Method
You are here!

Partial and Sequential Screening of Genes
Some of the tests offered by the Carver Laboratory are based upon partial screening of the coding sequences of one or more disease-causing genes. Moreover, the tests are often performed in a tiered fashion, with the portions that are most likely to yield results done first. This allows us to find clinically meaningful sequence variations as inexpensively as possible.
The LCA Example
To understand why this is so, consider the case of an infant with a recent clinical diagnosis of the autosomal recessive condition known as Leber congenital amaurosis (LCA). Her parents would like a molecular diagnosis right away to confirm the clinical impression of LCA, and they are asking their genetic counselor whether it will be possible at some point for them to have preimplantation diagnosis to avoid having a second affected child. Although they would never phrase it in this way, the genomic challenge from their point of view is: “How can you find the mutations that are responsible for my child's disease as rapidly and inexpensively as possible?”
In 2003, there are six genes that have been convincingly shown to cause the LCA phenotype in humans: Guanylate Cyclase, RPE65, RPGRIP, CRX, CRB1, and AIPL1 [5-11]. A screen of the entire coding region of these six genes by either automated DNA sequencing or single-strand conformation polymorphism analysis (SSCP) requires the study of 108 different PCR products (amplimers). How should one proceed to get the answer that this family wants as rapidly and inexpensively as possible? Should the genes be screened one at a time, or all six at once? If screened sequentially, which one first? The largest? The smallest? In alphabetical order? Should the entire coding sequence of every gene be screened?
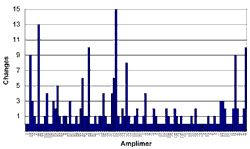
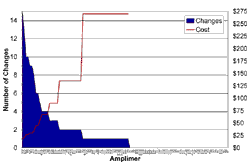
Figures 1 and 2 are interesting to consider in the context of these questions. Figure 1 shows all of the sequence variations with a reasonable likelihood of causing disease (i.e., with an Estimate of Pathogenic Probability score of 2 or 3) that we detected in the coding sequences of these six genes in a cohort of over 300 LCA probands (the mutations themselves are tabulated elsewhere on this website). In Figure 1, the genes are arranged in alphabetical order and within each gene the amplimers are arranged as they are in the genomic sequence with the 5' end of the gene to the left. The value depicted on the Y axis is the total number of potentially disease-causing sequence variations found in each individual amplimer during the screen of the entire cohort. Figure 2 shows the same data displayed in a different way. In this case, the amplimers have been ranked from left to right according to the number of potentially disease-causing variants that they were found to contain. That is, the amplimer containing the highest number of total variations (13) is placed at the left and the 32 amplimers that did not harbor a single variation are placed to the right.
Assuming that the new patient in this hypothetical example was drawn from a similar population as the cohort that we have already screened, each Y axis value (mutations per amplimer) of Figure 2 is proportional to the probability of finding one of the patient's two mutations in the amplimer with that Y value. Careful inspection of this figure shows that the ten amplimers that are most likely to contain a mutation are nearly equally distributed among four different genes. Also, one can see that these 10 amplimers (which comprise only 8 percent of the total coding sequence of these six genes) contain nearly half of all the mutations that we have found to date. By sorting the amplimers according to the probability that they will contain this patient's mutation, it allows one the opportunity of discovering 50 percent of all there is to find with only 10 percent of the effort of a "complete screen."
For those who feel a compulsion to "be complete" it is important to recognize the unfortunate truth that screening the entire coding sequences of these six genes by SSCP does not in any way represent a truly complete mutation screen of all LCA genes. To begin with, these genes are collectively responsible for only about a third of all cases of LCA [11], and even within these genes, there are undoubtedly mutations that exist in the promoter and other non-coding regions that would not be detected by a PCR-based assay of the coding sequence.
The red line in Figure 2 depicts another meaningful value, the cost of finding each mutation in each amplimer. This is obtained by dividing the cost of screening the entire cohort with each amplimer, by the number of mutations found in that amplimer. Looking at the data in this way, one can see that it costs very little to find mutations in the first 10 amplimers (at the left of the figure) while the cost per mutation is so high that we cannot even estimate it for the amplimers to the right. Suppose the parents are willing to pay $500 for the 17.5 percent chance of finding a discoverable mutation in the first 10 amplimers (i.e., 50 percent of the 35 percent maximum chance of finding an LCA mutation with our current level of knowledge). Would they be willing to pay $5000 for the remaining 17.5 percent opportunity that is present in the next 50 amplimers? Would they be willing to pay another $5000 for the less than 1 percent chance in the remaining 40 amplimers? Should every patient be screened on every amplimer even if two high probability mutations (EPP of 2 or 3) have been found on separate alleles of the same gene early in the screening process? That is, just to make sure that nothing is missed, should every patient have a $10,000 test? After six more LCA genes of equal size have been discovered, should every patient have a $20,000 test just to make sure that nothing is missed? Probably not. We believe that to maximize the benefit derived from genetic testing, we should tailor the tests that we perform to the specific research or clinical situation.
Summary
To summarize the main points made in this section:
- 100 percent diagnostic certainty is not attainable with this type of testing; and,
- trying to attain 100 percent diagnostic certainty for large numbers of people will be prohibitively expensive and time consuming.
« Return to the Genetic Testing FAQs